R3DAlign Help
About
The web server implements R3D Align (‘RNA 3D Align’) for global, locally optimized, nucleotide to nucleotide pairwise alignments of RNA 3D structures. R3D Align constructs a global alignment by examining local superpositions to produce a highly accurate alignment. The web server features an intuitive interface and highly detailed forms of output for the user. The website also provides the first of its kind “Gallery of Featured Alignments”, which includes R3D Align alignments of 5S, 16S, and 23S ribosomal RNA 3D structures of various organisms. The method is described by Rahrig et al., 2010.
A full description of how to effectively use R3D Align and the full features it offers are explained below.
Input Page and Settings
The only required input is the specification of the two RNA 3D structures to align. The user may upload a structure or enter a valid PDB id. As the user types in the PDB id, an auto-complete function assists the user in entering the proper id by providing a list of PDB ids that begin with the already-typed characters:
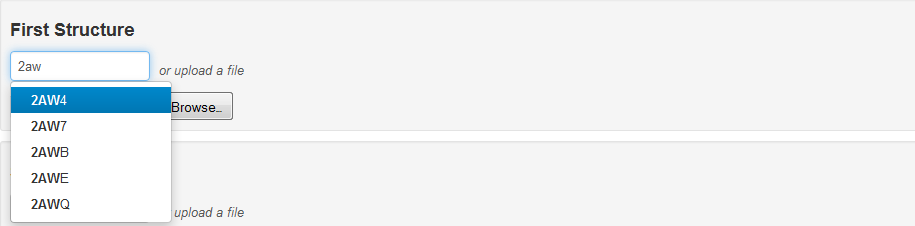
The list of files to choose from is updated weekly to be in sync with the files available from the PDB database.
Once the structure is selected from the list, the chains contained in the file are dynamically loaded, with their lengths and brief descriptions, for the user to choose from:
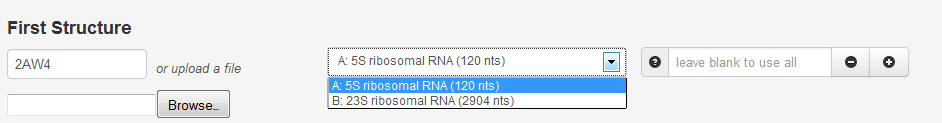
In the box on the right, specific nucleotides can be entered to align only a fragment of a chain or it can be left blank include all nucleotides in the chain. A range of nucleotides can be entered using a colon, and multiple ranges can be entered separated by a comma, such as "2:20, 60:69, 110:119". Hovering over the question mark icon reminds the user of this format. Clicking the plus sign icon allows the user to select another chain (and/or specific fragment, if desired) to be included in the alignment.
Once the two structures and nucleotides have been selected, the user may choose to use the default alignment parameters and hit the submit button in the lower right corner:
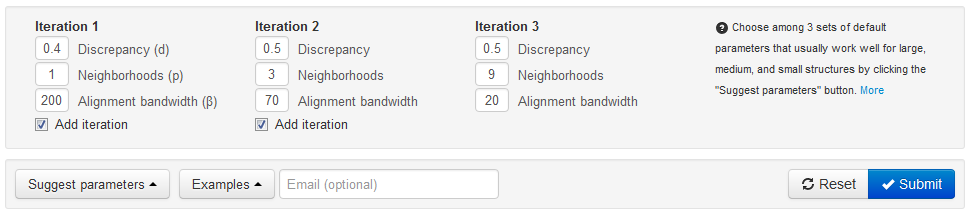
The default parameters have been optimized through a series of rigorous tests of thousands of alignments. The default parameters are suitable for structures of all sizes; however, additional Suggested Parameters based on the size of the structures are available by clicking the ‘Suggest Parameters’ button in the lower right hand corner.
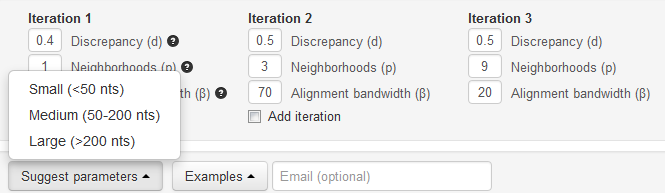
Using these parameters will retain the quality of the alignment that would be obtained using the default values, but will decrease the run time required to produce the alignment. Since the suggested parameters are based on the size of the structures, it is simple for the user to choose the appropriate parameters. Once a structure size is selected, the default parameters are dynamically loaded into the alignment parameters.
Clicking the examples button provides a list of examples which, if selected, will automatically load into the input page for submission. Hitting submit will immediately take the user to the results page for the selected alignment. One can also learn about good alignment strategies by examining the parameters used in the Gallery of Featured Alignments.
Iterative alignment
The R3D Align web server has been designed to allow alignments to be done in an iterative fashion. For medium to large structures (more than 150 nucleotides), using an iterative approach typically produces more accurate alignments in a shorter period of time. With an iterative approach, the alignment produced by one iteration is used as a seed alignment for the subsequent iteration. R3D Align uses an iterative approach by default.
In general, the user should increase the value of p from one iteration to the next while decreasing the value of β. Setting a low value of p (1-2) in the first iteration limits the number of four-nucleotide neighborhoods considered by R3D Align and produces an alignment quickly. A large bandwidth β (100-200) should be used for this first iteration since an internally-generated sequence alignment is used as the seed. The resulting alignment, which is much more accurate than the seed alignment used in the first iteration, is used as the seed alignment for the second iteration. Because this seed alignment is more accurate, a smaller bandwidthβ (50-100) can be used for the second iteration, which decreases runtime and allows a higher value of p (4-6) to be used to produce an even more accurate alignment. This more accurate alignment can be used as a seed for a third iteration for which a low value ofβ (<25) can be used and a large value of p (8-10). This iterative approach produces results similar to simply running one iteration with large values of p and β, but in significantly less time.
The next section describes how to adjust the alignment parameters for the purpose of tweaking the results produced by the R3D Align algorithm.
Advanced Alignment Parameters d, p, and β
- Discrepancy cutoff (d)
Geometric discrepancy is a measure of how similar two RNA (sub)structures are. Identical structures have discrepancy zero. Higher geometric discrepancy corresponds to more dissimilar structures. R3D Align works by identifying pairs of four-nucleotide neighborhoods, one from each structure, which have geometric discrepancy lower than the discrepancy cutoff d. Typical values of dlie between 0.3 and 0.6. Higher cutoff values result in aligned regions that are less geometrically similar, but are more tolerant of structural differences between structures. Alignments with high geometric discrepancy cutoffs take longer to produce than those with lower cutoffs because more pairs of four-nucleotide neighborhoods are generated. For more details on the development of the discrepancy measure, click here.
- Number of neighborhoods per nucleotide (p)
The parameter p indicates the number of four-nucleotide local neighborhoods that will include each nucleotide. For each nucleotide in each structure, the p neighborhoods with the smallest diameter (maximum distance between any two nucleotides) will be compared with the local neighborhoods in the other structure. Typical values of p fall in the range from 1 to 10.
Larger values of p provide more coverage for each nucleotide, which typically improves the accuracy of the alignment. However, increasing the value of p also has the effect of increasing the running time of the program, often significantly. When aligning small regions or small structures, this increase is not significant. When aligning medium to large regions, it is suggested to use an iterative approach in which a smaller value of p (1-2) is used in the first iteration, a moderate value of p (4-6) is used in the second iteration, and a large value of p (8-10) is used in the third iteration. The progressively increasing values of p are offset by the progressively decreasing values of the bandwidth parameter β (described below) to maintain a reasonable run time.
- Bandwidth (β)
The bandwidth parameter (β) is an option that can be used to improve runtime. β is a positive integer that indicates how far away from the internally-generated seed alignment R3D Align should look for its local geometric alignment. It is set according to how accurate the seed alignment is believed to be. If a seed alignment aligns nucleotide i of Molecule 1 with nucleotide j of Molecule 2, then only alignments in which nucleotide i is aligned with a nucleotide in the range j-β/2 to j+β/2 will be considered (β/2 is rounded to the nearest integer). This reduces the search space, which decreases runtime.
For the first iteration of an alignment an internally produced sequence alignment is used as the seed alignment. β should be set according to how accurate a sequence alignment is believed to be (overestimate if unsure). Set β high for the first iteration if a sequence alignment is not expected to be close at all to the actual alignment (or just to be safe). Setting β larger than necessary does not have much of an effect on the accuracy of the alignment, although it does increase the time required to compute the alignment.
The bandwidth entered for the second or third iteration is applied to the alignment produced by R3D Align in the previous iteration in the same way that the bandwidth entered for the first iteration is applied to the seed alignment. In subsequent iterations, the alignment determined by R3D Align in the previous iteration is used as the seed alignment. If one is progressively raising p (as described above) in an iterative alignment, the alignments will get progressively better. This allows for a progressive decrease in bandwidth (β) and a control of the computational time.
To consider all possible alignments, set β greater than or equal to twice the number of nucleotides to be aligned for the second structure. This has the effect of ignoring the seed alignment.
Query Submission/Processing Page
The R3D Align web server implements extensive pre-submission validation to ensure that user submitted input can be processed. This includes validation of specified nucleotides to prevent the server from attempting queries containing non-existent nucleotides.
Notably, in the instance that a user submits a query that has previously been processed, the user is taken immediately to the results page without waiting for processing as the results are retrieved from a database. Each such submission generates a new randomly-generated URL.
In other cases, upon submission the user is redirected to an interstitial page that can bookmarked for future reference, similar to the one below:
If a job has not begun to be processed, the line "Your job request is being processed" will appear as "Your job request has been successfully submittted".
Clicking the Share button allows the user to bookmark and share the page via such options as e-mail, Gmail, Facebook, Twitter, etc.
Results Page
The R3D Align results page provides the user with a wealth of information regarding the computed alignment in a variety of formats. The results are stored on the server indefinitely with stable URLs, which makes it easy for collaborators to share results. URLs include a long randomly-generated text string, revealed only to the user, to protect the privacy and security of user data and analyses. The following figure illustrates the form of a R3D Align results page:
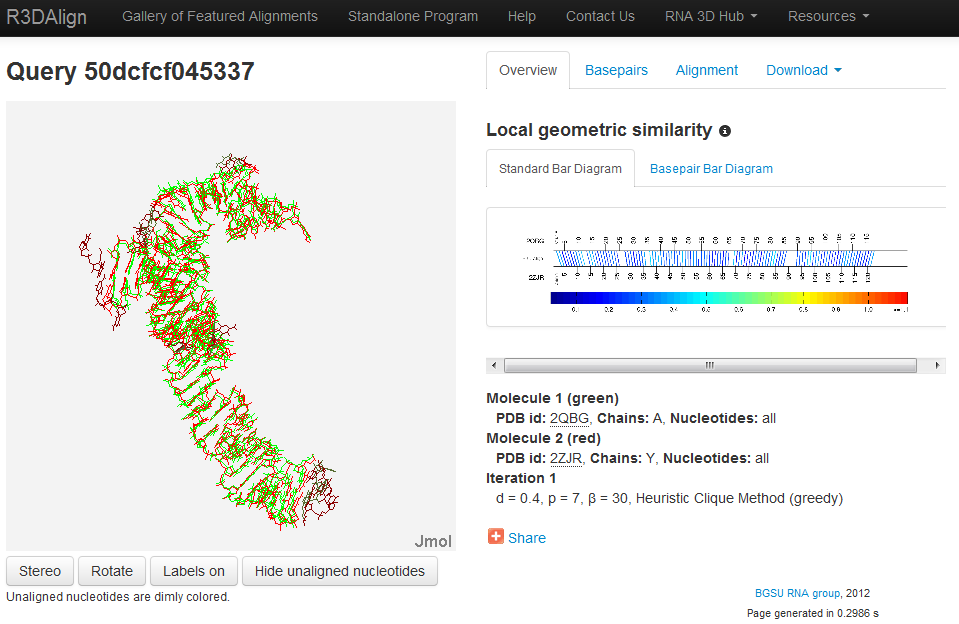
Jmol Display
On the left side of the screen is an interactive Jmol applet displaying a rigid superposition of the two structures. Note that a rigid superposition necessarily only approximates the local nature of the nucleotide to nucleotide alignment provided by R3D Align. Nucleotides in the first structure are colored green and those from the second structure are colored red. Nucleotides found to have a corresponding aligned nucleotide by R3D Align are brightly colored.
The right side of the screen features summary output of the alignment in a variety of formats including numerical, textual, and visual. The user can toggle between the different displays using the tabs at the top. All output is directly accessible on the results page for viewing and interaction, although files can be downloaded for local use by clicking Download.
We created a brief tutorial to demonstrate how to understand R3D Align results using Jmol.
Overview and Bar Diagram Representation
The overview tab provides the user with summary statistics of the alignment in addition to a review of the parameters used for the alignment procedure. This tab also displays the Standard Bar Diagram with an option to toggle to view the enhanced Basepair Bar Diagram by clicking the Basepair Bar Diagram tab. Clicking on the bar diagram enlarges the image for the user. For more information on the bar diagrams and how to interpret them see the R3DAlign Bar Diagram Help.
Basepair Table Representation
Clicking the Basepairs tab displays the alignment in tabular form, illustrating the alignment of basepairs in the two structures. Like the bar diagram this display offers a quick, yet highly detailed visual assessment of the alignment. The following is an example of the display when this option is selected:
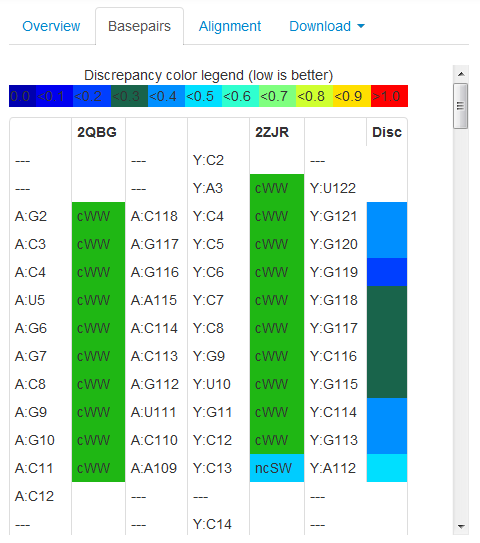
For more information on this output display and how use it properly, see the R3D Align Basepair Table Help.
Text Alignment Representation
The Alignment tab provides the user with a summary of the alignment in text format, alternately listing the nucleotides from Structure 1 and Structure 2 in FASTA format:
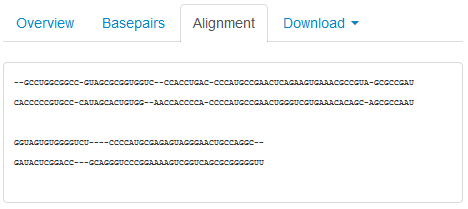
Downloading Files
All output files are downloadable for local use. Upon clicking the Download tab, the user is presented with the following options:
Updated: 08/01/2025 05:42PM
