Announcements
This week we are rolling out an update to WebFR3D, which includes several enhancements:
1. Seamless integration with FR3D
In the past upgrading FR3D, the computational engine powering WebFR3D, was done fairly rarely, and as a result improvements in FR3D basepair annotations and search algorithms were not making their way to WebFR3D. Now the official FR3D repository is included in WebFR3D as a submodule, which allows for much better control over what version of the software is deployed.
2. Integration with RNA 3D Hub
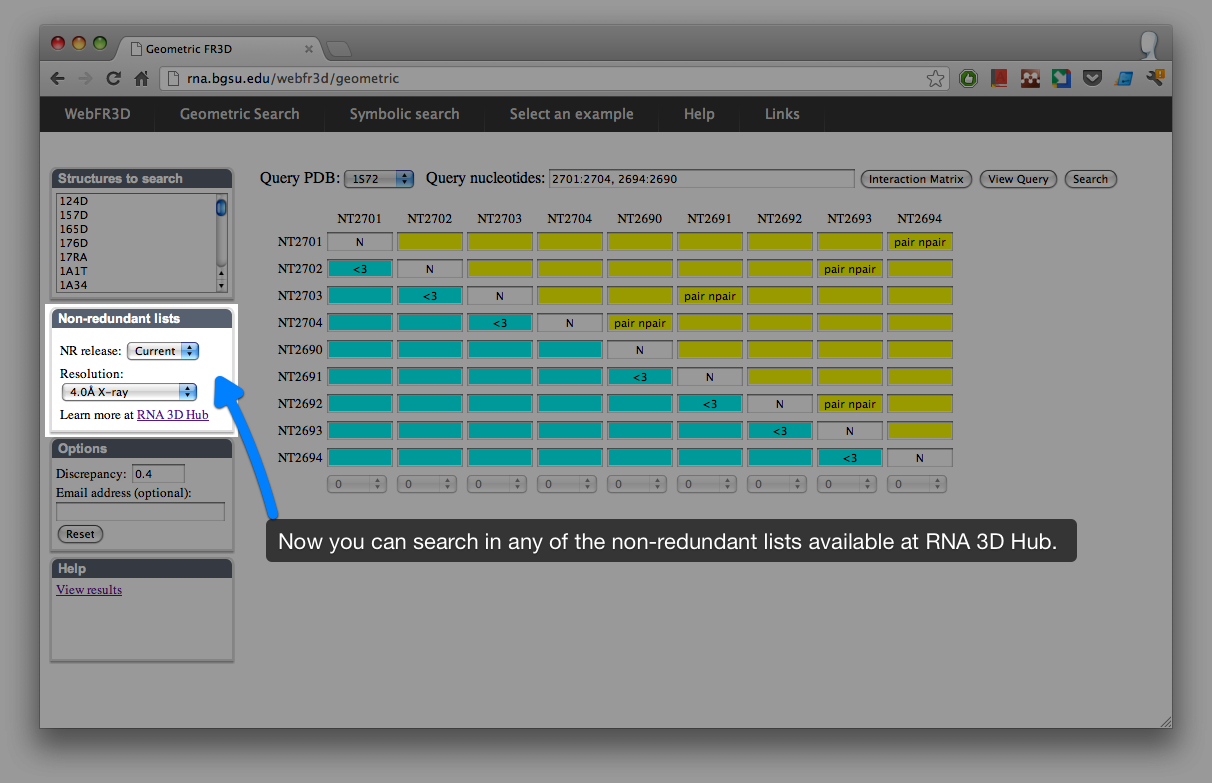
Since the beginning it was possible to use WebFR3D to search non-redundant lists of RNA 3D structures. However, these lists had to be updated manually and this wasn’t done regularly. Now WebFR3D is integrated with the Non-redundant Atlas, which is a part of RNA 3D Hub. The non-redundant lists are automatically updated every week.
3. Other improvements
- Bug fixes related to sending email notifications.
- WebFR3D informational website has been integrated with the rest of the rna.bgsu.edu website.
- All old WebFR3D results should still be available.
- WebFR3D code is now hosted on Github.
Please let us know if you encounter any problems with WebFR3D by using the contact form, by submitting an issue on Github, or by tweeting at RNA 3D Hub.
- WebFR3D paper is online April 7, 2011Anton Petrov
WebFR3D paper is accepted for publication in the 2011 Webserver Special Issue of Nucleic Acids Research. You can get early access here.
WebFR3D development is continuing with more features coming soon. Let us know what you think about WebFR3D and what is on your wishlist using the contact form.
- More search examples, minor interface tweaks March 7, 2011Anton Petrov
We have added more examples of geometric WebFR3D searches; now they include sarcin-ricin, triple-sheared, kink-turn, C-loop and T-loop. You can view the search parameters and launch a new search, or examine the precomputed results by clicking on ‘View results’ link in the Help section on the bottom left of the screen.
Fixed an issue with the Jmol applet on the results page overlaying the PDB info popup window. Thank you for pointing this problem out!
- WebFR3D is in sync with PDB February 7, 2011Anton Petrov
The RNA-containing PDB files are downloaded from PDB on a weekly basis. The new structures should become available for searching in WebFR3D on Sunday of the week when the structures are released. Automatic updating of the nonredundant lists is coming soon.
- Redesigned WebFR3D is here! Here is a list of new features: December 7, 2010Anton Petrov
- Improved validation of user input. All nucleotides and all chains are checked before the user submits the form, so that non-existent nucleotides cannot be used in queries.
- The user can preview the query in Geometric mode. The preview window is draggable. Nucleotide numbers can be turned on and off, and the pdb file can be downloaded. This can be used as a tool for quickly extracting a short fragment from a pdb file.
- Contextual help added – when the user clicks an element on the webpages, brief help messages appear in the designated Help section of the page. If that information is not sufficient, the user can click “More” to see more help in the overlayed window, which doesn’t force the user to leave the page.
- More example searches are added. The user can view precomputed results by clicking ‘View results’ in the Help section of the page.
- Enhanced output webpages. The interaction annotations now include base phosphate interactions. Empty columns are no longer displayed. The annotations table is visualized with the Tablecloth javascript library.
- Added 16 Angstrom neighborhood to result structures.
- Next/previous and Lock functions for jmol visualization. Keyboard shortcuts added: j to view previous structure, k to view the next one.
Updated: 10/22/2025 10:09AM
