Accessing RNA 3D Hub Data
FR3D Pairwise Interaction Annotations
Downloading all interaction annotations at once
A gzipped csv file with FR3D interactions can be downloaded at the following url: http://rna.bgsu.edu/rna3dhub/data/interactions.gz (as of 1/1/2020 it was 175MB compressed, 1.5GB uncompressed).
The file looks like this (the format is specified in the header line):
"unit_id1","unit_id2","FR3D basepair (f_lwbp)","FR3D stacking (f_stacks)","FR3D base phosphate (f_bphs)"
"124D|1|B|A|10","124D|1|B|C|9","","s53",""
"124D|1|B|C|9","124D|1|B|A|10","","s35",""
…
"8PSH|1|B|G|15","8PSH|1|B|G|15","","","0BPh"
"8PSH|1|B|C|16","8PSH|1|B|C|16","","","0BPh"
The interactions are listed using new-style unit ids. At present, PDB files for which new-style unit ids are not available yet (for example, viral 3D structures), do not appear in this file.
The interactions.gz file is updated on Wednesday mornings as part of the RNA 3D Hub update.
Downloading interaction annotations for individual PDB files
FR3D annotations can also be downloaded for individual PDB files using web interface or by accessing a url formatted as follows:
http://rna.bgsu.edu/rna3dhub/pdb/<pdb_id>/interactions/fr3d/<interaction_type>/csv
<pdb_id> is a case-insensitive four-character pdb identifier.
<interaction_type> is one of the following:
basepairs
stacking
basephosphate
baseribose
all
Examples: 1J5E basepairs, stacking, basephosphate, baseribose, all interactions in one file.
Non-redundant Lists
Non-redundant lists can be downloaded manually using the download tab:
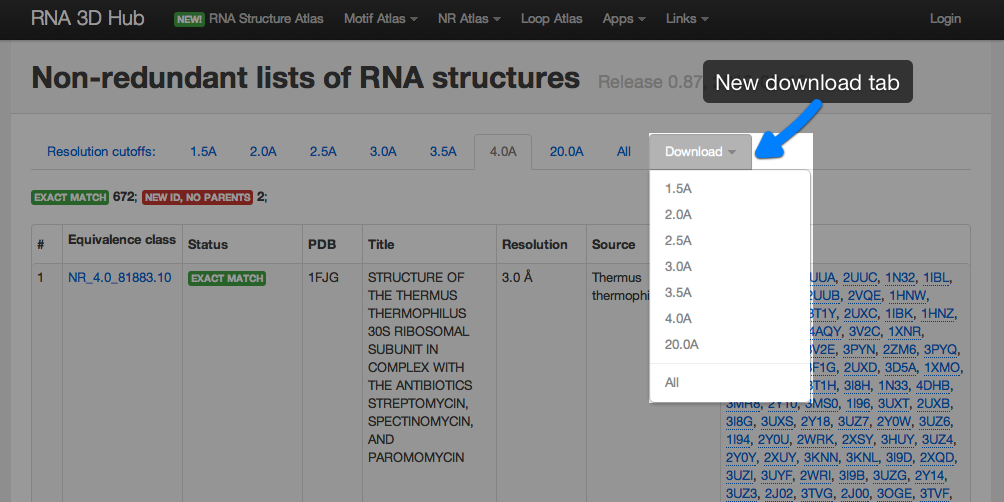
The current non-redundant lists can be accessed programmatically at a standard url:
http://rna.bgsu.edu/rna3dhub/nrlist/download/current/<resolution_cutoff>/csv
where <resolution_cutoff> is one of the following:
1.5A (1.5A – 4.0A for X-ray structures only)
2.0A
2.5A
3.0A
3.5A
4.0A
20.0A (X-ray + cryoEM)
all (X-ray + cryoEM + NMR structures)
The data are available in the following format:
"Equivalence class id","Representative structure","Comma-separated list of the equivalence class members"
For example, http://rna.bgsu.edu/rna3dhub/nrlist/download/current/1.5A/csv:
“NR_1.5_23181.1″,”2ASB”,”2ASB”
“NR_1.5_11995.1″,”3HGA”,”3HGA”
“NR_1.5_76588.1″,”397D”,”397D”
“NR_1.5_49442.1″,”2R22″,”2R22″
“NR_1.5_27899.1″,”2Y8Y”,”2Y8Y”
“NR_1.5_93883.1″,”3ND3″,”3ND3,3ND4″
This sample Python script can be used for downloading the current non-redundant lists.
Any non-redundant list can be downloaded using the following URL format:
http://rna.bgsu.edu/rna3dhub/nrlist/download/<release_id>/<resolution>/csv
For example: http://rna.bgsu.edu/rna3dhub/nrlist/download/0.85/1.5A/csv
RNA 3D Motif Atlas
Downloading individual entries
Each entry can be downloaded manually:
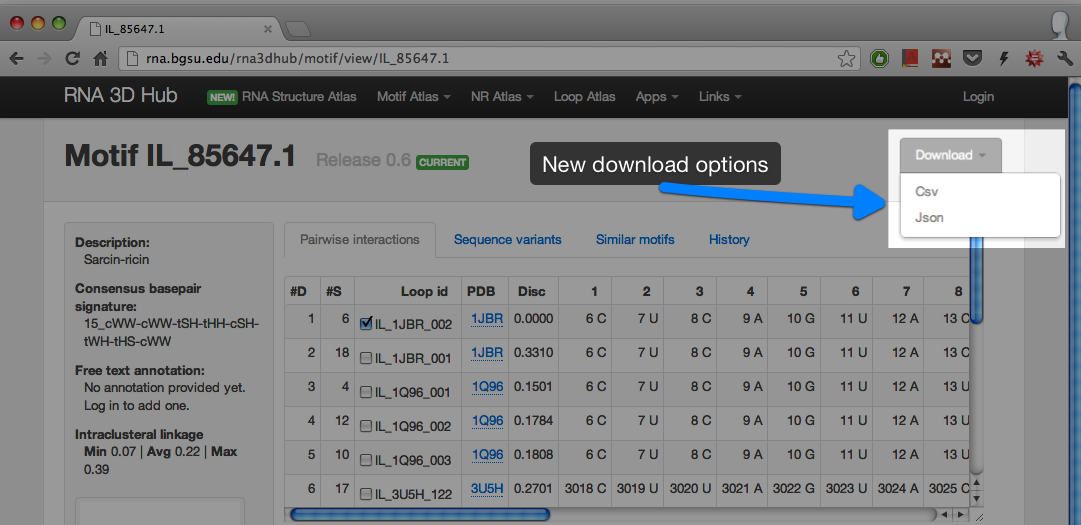
The data are available in two formats:
csv
Each line represents a motif instance and shows the aligned new-style unit ids of its constituent nucleotides. Here is an alignment of 2 motif instances:
"3UCZ|1|R|G|76","3UCZ|1|R|U|77","3UCZ|1|R|A|61","3UCZ|1|R|A|62","3UCZ|1|R|A|63","3UCZ|1|R|C|64"
"3IWN|1|A|G|71","3IWN|1|A|U|72","3IWN|1|A|A|51","3IWN|1|A|A|52","3IWN|1|A|A|53","3IWN|1|A|C|54"
json
Json can be easily parsed in most programming languages and is convenient because it provides the data in a self-documented way. This sample Python script illustrates the current API implementation (requires python > 2.6). Here is an example json document:
{
"num_instances":16,
"num_nucleotides":15,
"alignment":{
"IL_1Q96_001":[
"1Q96|1|A|C|6",
"1Q96|1|A|U|7",
"1Q96|1|A|C|8",
"1Q96|1|A|A|9",
"1Q96|1|A|G|10",
"1Q96|1|A|U|11",
"1Q96|1|A|A|12",
"1Q96|1|A|U|13",
"1Q96|1|A|A|18",
"1Q96|1|A|G|19",
"1Q96|1|A|A|20",
"1Q96|1|A|A|21",
"1Q96|1|A|C|22",
"1Q96|1|A|C|23",
"1Q96|1|A|G|24"
],
(some entries are left out for demonstration purposes)
"IL_2ZJR_045":[
"2ZJR|1|X|A|1275",
"2ZJR|1|X|U|1276",
"2ZJR|1|X|G|1277",
"2ZJR|1|X|A|1278",
"2ZJR|1|X|G|1279",
"2ZJR|1|X|U|1280",
"2ZJR|1|X|A|1281",
"2ZJR|1|X|A|1282",
"2ZJR|1|X|U|1994",
"2ZJR|1|X|G|1995",
"2ZJR|1|X|A|1996",
"2ZJR|1|X|A|1997",
"2ZJR|1|X|A|1998",
"2ZJR|1|X|U|1999",
"2ZJR|1|X|U|2000"
]
},
"motif_id":"IL_85647.2",
"common_name":"Sarcin-ricin parent motif with 15 Nts",
"annotation":"Complete Sarcin-ricin IL motif",
"bp_signature":"cWW-cWW-tSH-tHH-cSH-tWH-tHS-cWW"
}
Downloading Motif Atlas releases
The most current internal loop release can be downloaded at the following urls (csv and json formats):
http://rna.bgsu.edu/rna3dhub/motifs/release/il/current/csv
http://rna.bgsu.edu/rna3dhub/motifs/release/il/current/json
To download hairpin loop releases, replace “il” with “hl”:
http://rna.bgsu.edu/rna3dhub/motifs/release/hl/current/csv
http://rna.bgsu.edu/rna3dhub/motifs/release/hl/current/json
All Motif Atlas releases are archived and can be downloaded independently by replacing “current” with release number (e.g. 0.7 or 1.0). For example: http://rna.bgsu.edu/rna3dhub/motifs/release/il/0.7/csv
Downloading loops for individual PDB files
Loops can be downloaded manually by clicking the Download button:
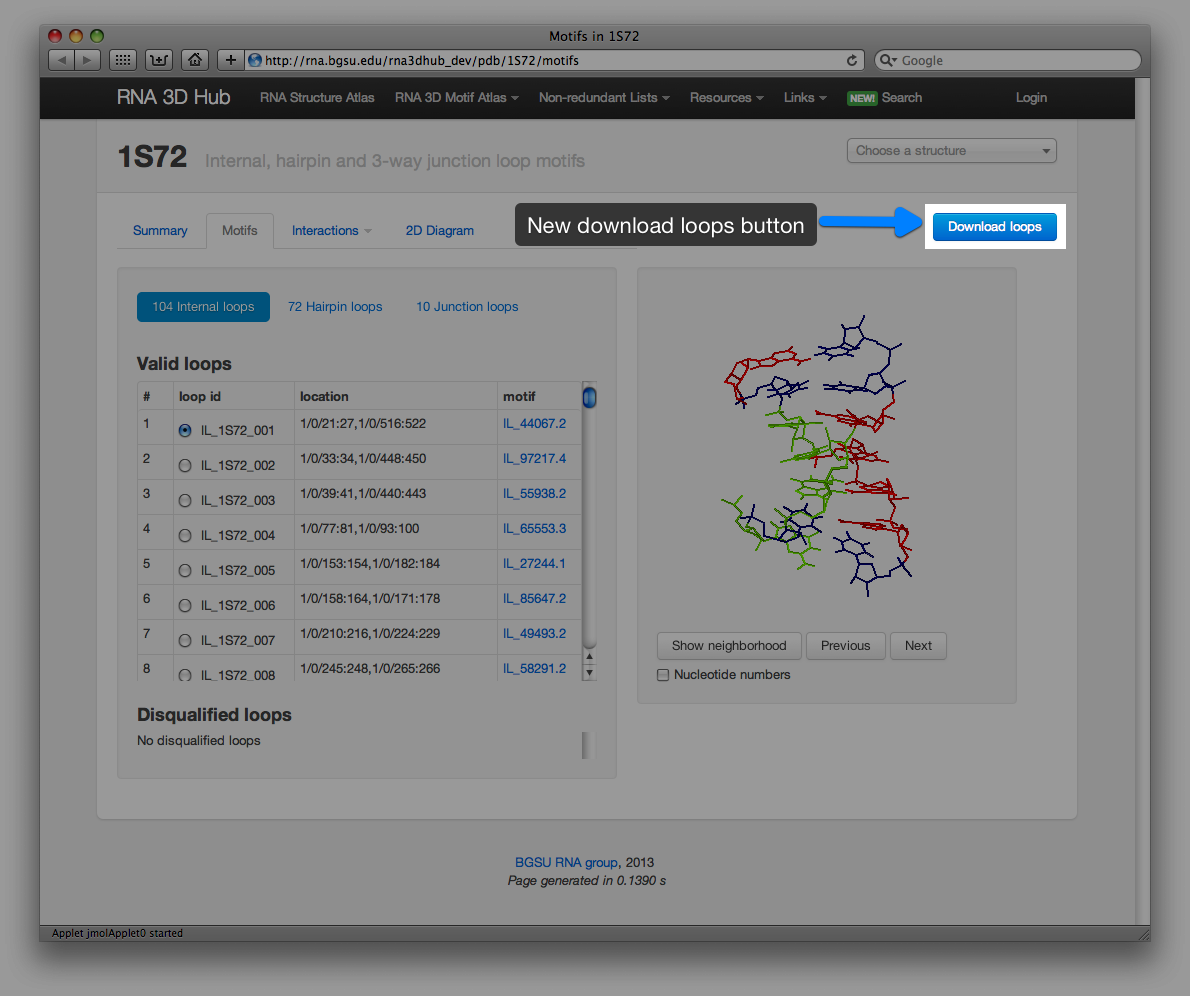
The files are in csv format:
"loop_id","comma-separated unit ids"
For example:
"HL_1S72_003","1S72|1|0|U|121,1S72|1|0|C|122,1S72|1|0|G|118,1S72|1|0|A|119,1S72|1|0|A|120"
"HL_1S72_004","1S72|1|0|U|125,1S72|1|0|A|128,1S72|1|0|A|129"
If the pdb file doesn’t exist or there are no loops, the file will say: “No loops found.”
One can also download the loops programmatically by accessing the URLs formatted as follows:
http://rna.bgsu.edu/rna3dhub/loops/download/<pdb_id>
For example: http://rna.bgsu.edu/rna3dhub/loops/download/1S72
Downloading All Loops
The current set of all extracted loops, excluding any loops from obsoleted files, can be downloaded at:
http://rna.bgsu.edu/rna3dhub/data/loops.csv.gz
This is a comma separated file with four columns: the loop id, assigned motif id if any, the pdb id for the loop and a comma separated list of the nucleotide ids. All fields are quoted with ” (double quotes). The file includes a header line. An example of the first few lines from the file is below:
"id","motif_id","pdb","nts"
"IL_157D_001","IL_47174.6","157D","157D|1|A|C|3,157D|1|A|G|4,157D|1|A|A|5,157D|1|B|U|20,157D|1|B|A|21,157D|1|B|G|22"
"IL_157D_002","IL_47174.6","157D","157D|1|A|G|10,157D|1|A|U|8,157D|1|A|A|9,157D|1|B|C|15,157D|1|B|G|16,157D|1|B|A|17"
"IL_165D_001","IL_02809.2","165D","165D|1|A|U|3,165D|1|A|U|4,165D|1|A|C|5,165D|1|A|G|6,165D|1|B|U|12,165D|1|B|U|13,165D|1|B|C|14,165D|1|B|G|15"
"IL_17RA_001","","17RA","17RA|1|A|C|15,17RA|1|A|U|16,17RA|1|A|A|17,17RA|1|A|U|5,17RA|1|A|A|6,17RA|1|A|A|7,17RA|1|A|G|8"
If a PDB file has no loops, it will not appear in this file. If the loop has not been assigned a motif the motif_id column will be “”, as seen in the last line of the example above.
Linking to RNA 3D Hub
Urls for the RNA 3D Hub pages dedicated to PDB files look like this:
http://rna.bgsu.edu/rna3dhub/pdb/1J5E
The four-character PDB id is case-insensitive.
Updated: 08/01/2025 05:42PM
