R3D-2-MSA Server
The R3D-2-MSA server makes it possible for users to access high-quality RNA multiple sequence alignments curated by Robin Gutell’s research group at the University of Texas at Austin to obtain sequence variants for RNA structural motifs observed in atomic-resolution RNA 3D structures. Users can simply use the nucleotide numbers from representative atomic-resolution RNA 3D structures from PDB/NDB to access the corresponding columns of the alignment. The server automatically makes the appropriate mappings. Currently, mappings are provided for small and large subunit (SSU and LSU) ribosomal RNA (rRNA) structures from bacteria (SSU and LSU), eukarya (SSU and LSU) and archaea (LSU). When a structure file contains more than one rRNA molecule, users can query ranges from different molecules, for example from 5S, 5.8S, and 26S rRNA in eukaryal LSU.
Input form for human users
Starting at the main R3D-2-MSA page the dropdown menu lists available combinations of RNA 3D structures and available alignments to query for sequence variations. First select a representative structure and then use the radio buttons to select the desired alignment, when more than one is available. Consult the examples to see how to enter nucleotide ranges for 1) two paired bases; 2) a hairpin loop; 3) an internal loop; or 4) a three-way junction. Note that in case (1), each range consists of a single nucleotide, and so no intervening columns are returned, but in cases (2), (3), and (4), each range will return the specified nucleotides and all intervening columns of the alignment.
Annotations of basepairs, other pairwise interactions, and internal and hairpin loops are available for each 3D structure file; look up the file on the NDB server or the BGSU server. These will aid in the selection of relevant nucleotide ranges.
URL input
Queries can be formed directly as a URL instead of using the input page. Here are the URL inputs corresponding to the four examples available from the R3D-2-MSA main page:
1) The URL for a query corresponding to two bases from E. coli SSU which make a Watson-Crick basepair.
http://rna.bgsu.edu/r3d-2-msa?units=2AW7|1|A||1265,2AW7|1|A||1270&aid=1
The comma separates the first and second base of the basepair, and no other columns of the alignment are returned. The nucleotides themselves are encoded using the nucleotide IDs we recently developed in consultation with NDB. For a description of nucleotide IDs please see this page.
2) The URL encoding a query for the 16S rRNA helix 41 GNRA hairpin loop is the following:
http://rna.bgsu.edu/r3d-2-msa?units=2AW7|1|A||1265:2AW7|1|A||1270&aid=1
The colon ( ‘:’ ) separates the starting and ending nucleotides of a single range of nucleotides. The R3D-2-MSA server will return the columns corresponding to these nucleotides and all intervening columns of the alignment, including columns present because of insertions in one or more sequences. Note that the base letter (A, C, G, or U) is not specified in the nucleotide unit ids in this example; they are not actually needed by the R3D-2-MSA server.
3) Here is the URL encoding a query for the internal loop in helix 20 of 16S rRNA:
Here the comma ( ‘,’ ) separates the two nucleotide ranges defining the internal loop. Up to five separate ranges can be input in a single query. Currently up to 50 nucleotides total can be queried at once.
4) This URL will return sequence variations of a three-way junction in bacterial 16S rRNA:
http://rna.bgsu.edu/r3d-2-msa?units=2AW7|1|A||826:2AW7|1|A||829,2AW7|1|A||857:2AW7|1|A||861,2AW7|1|A||868:2AW7|1|A||874&aid=1
Output page for human users
At the top of the output under “Query Information” a URL is provided that encodes the query that was carried out. This URL can be saved if desired to allow the user to repeat the search by simply pasting it into a browser, or the link can be embedded in other web pages to repeat the search. Search results are not saved on the server.

The next section of the output summarizes the distinct sequences found in the columns of the multiple sequence alignment corresponding to the query. Below is the output for query (1) above:
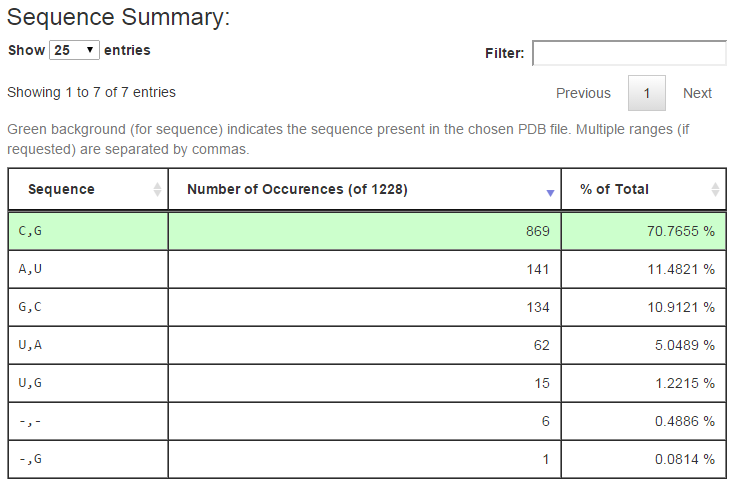
The first column of the table shows the distinct sequences retrieved from the alignment, with separate ranges separated by columns as in the URL input. The second column tallies the number of rows in the alignment having the given sequence, and the third column reports the percentage of all alignment rows this number represents. The row of the alignment, if any, which has the same sequence as is found in the associated 3D structure is shown with a green background. The controls above the table make it easy to change the number of rows show. Typing text in the Filter box restricts the rows shown to those having the given text.
The last section of the output page lists all sequences retrieved from rows of the alignment along with information about the source of the sequence. Below are a few rows from the Sequence Details section from example query (3), an internal loop from the bacterial 16S SSU rRNA.
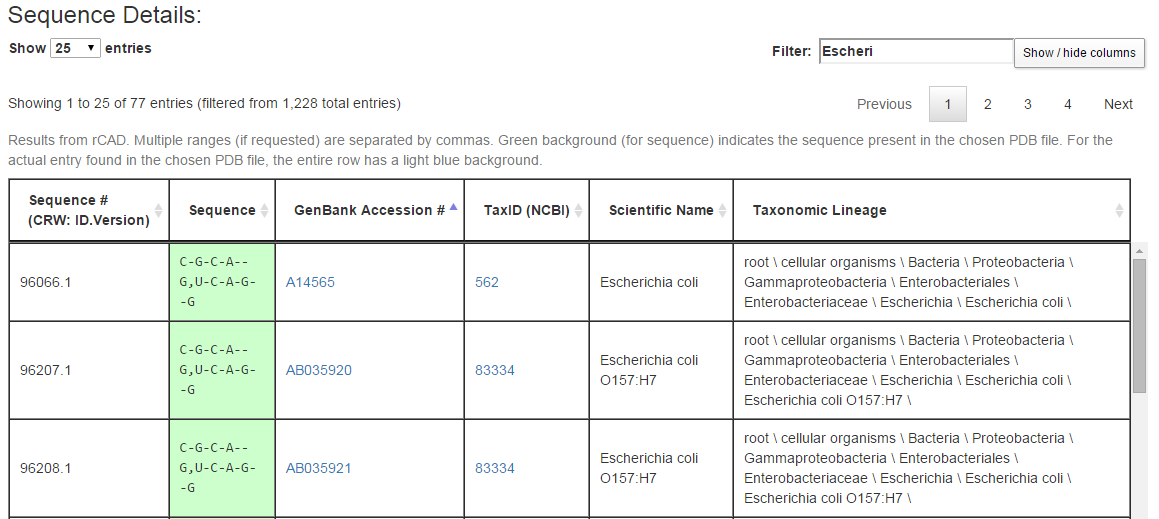
Each row of the Sequence Details table corresponds to a row from the original alignment. For the query above, that alignment has 1228 rows, of which three are shown in the figure above. The columns in each row show, in this order, an internal CRW identifier for the biological source for the row of the alignment, the portion of the sequence corresponding to the current query, the GenBank accession number of the biological source of the full sequence, the NCBI TaxID for this biological source, the scientific name, and the full NCBI taxonomic string of the biological source. The user can suppress the display of one or more columns using the “Show / hide columns” button. As in the Sequence Summary, green coloring is used for cells with the same sequence as in the 3D structure used to form the query. In addition, the row corresponding to the organism whose 3D structure was determined is colored with a blue background.
The “Filter” box restricts the rows being displayed to those containing, somewhere in the row, the text typed in the box. In the figure above, the first seven letters of “Escherichia coli” are typed in, which restricts the rows being displayed to those 77 rows containing the text “Escheri,” in particular those from species Escherichia coli. By making judicious use of the Filter box, one can explore the sequence conservation across phylogenetic groups. For example, in example query (3), the sequence C-G-C-A–G,U-C-A-G–G is conserved across species Escherichia coli and genus Escherichia, nearly conserved over family Enterobacteriaceae and order Enterobacteriales, over 50% conserved in class Gammaproteobacteria, but less than 50% conserved across phylum Proteobacteria. Filtering and then sorting by sequence is particularly helpful.
Programmatic input and output
For programmatic access to sequence variations we suggest using python scripts to make requests. It is suggested to use the python requests package available at this link:
http://docs.python-requests.org/en/latest/
To install the python requests package simply type the following command at the command line:
pip install requests
Examples of requests are provide by the python code at the following gist:
import time
import requests
FINISHED = set(['succeeded', 'failed'])
# Some large and slow requests may fail because of the retry limit in requests.
# Set the retry higher for these cases.
# 3 seconds is a pretty good wait most requests should be done in < 10 seconds.
def fetch(*args, **kwargs):
response = requests.get(*args, **kwargs)
response.raise_for_status()
data = response.json()
while data['status'] not in FINISHED:
time.sleep(3)
response = requests.get(*args, **kwargs)
data = response.json()
return response
# Returned content type may be set by changing the accept
headers = {'Accept': 'application/json'}
# Access variations for one nucleotide
response = fetch("http://rna.bgsu.edu/r3d-2-msa",
params={'units': '2AW7|1|A|A|887'}, headers=headers)
data = response.json()
# Access variations while omitting the base letter
response = fetch("http://rna.bgsu.edu/r3d-2-msa",
params={'units': '2AW7|1|A||887'}, headers=headers)
data = response.json()
# Access variations for a range of nucleotides
# We can also use a format parameter to set the returned content type
response = fetch("http://rna.bgsu.edu/r3d-2-msa",
params={'units': '2AW7|1|A|A|887:2AW7|1|A|A|895', 'format': 'json'})
data = response.json()
# Access variations for multiple ranges
response = fetch("http://rna.bgsu.edu/r3d-2-msa",
params={'units': '2AW7|1|A|A|887:2AW7|1|A|A|895,2AW7|1|A|G|200:2AW7|1|A|C|210', 'format': 'json'})
data = response.json()
# It is possible to have up to 5 ranges in the list of ranges
View raw - https://gist.github.com/blakesweeney/568f542f80f1bcd14788
The output is a JSON document with the structure:
{
"pdb": "2AW7", // The PDB id for the requested range.
"model": 1, // The model number for the requested range.
"reqs": [], // The sequence for the requested collection of units
"summ": [], // A summary of the sequence variation
"full": [] // All observed sequence variation
}
The reqs object gives the sequence of the given range of units as well as summarizing how often that sequence is found in the alignment.
[
{
"SeqVersion": 1,
"SeqID": 96080,
"CompleteFragement": "A-G--UU-U-G-A-U--CA-U",
"TotalCount": 1228
}
]
summ is a summary of the observed sequence variation. It is a list with one entry per unique sequence. It has the structure:
[
{
"CompleteFragement": "A-G--UU-U-G-A-U--UC--",
"NumberOfAppearances": 1
}
]
CompleteFragement is the sequence and NumberOfAppearances is the count. For requests that have helpal ranges the sequence of each range will be separated by a comma.
The full object contains information about each sequence in the alignment. It gives the sequence of the given slice of the alignment as well as some metadata about the sequence. It has the structure:
[
{
"AccessionID": "X74066",
"TaxID": 435,
"SeqVersion": 1,
"ScientificName": "Acetobacter aceti",
"LineageName": "root \\ cellular organisms \\ Bacteria ...", // Truncated
"CompleteFragment": "A-G--UU-U-G-A-U--UC-U"
}
]
The AccessionID is the Genbank accession id for the sequence. TaxID is the NCBI taxon id for this sequence. SeqVersion is the Genbank version for this sequence. ScientificName is the full scientific name for the organism this sequence comes from. LineageName is the full taxonomic lineage for the sequence. CompleteFragement is the sequence observed.
Updated: 08/01/2025 05:42PM
